Technical and methodological development for state-of-the-art proteomics
Sample processing is crucial for sensitive and comprehensive proteomic analyses, especially prior to mass spectrometric analysis in LC-MS/MS systems. Here we have developed and adapted a wide range of sophisticated protocols. Their elaborate use enables us to characterize proteomes of different microbes, both for our independent research and in cooperation with numerous national and international partners.
Some examples are:
Purification and enrichment of extremely diluted proteome samples
We have succeeded in establishing a protocol for enrichment of highly dilute samples, including proteins found in body fluids and excreted into growth media. It is based on commercially available affinity beads that have to be conditioned for this purpose in a proper fashion. Additionally most sensitive purification of proteome samples is possible, e.g. to clean up samples from salts or nucleic acids, that would affect following mass spectrometrical measurement. As a striking feature, this purification protocol further allows for shipping/ sharing proteome samples without elaborate cooling logistics.
- Bonn et al. PMID: 24987932.
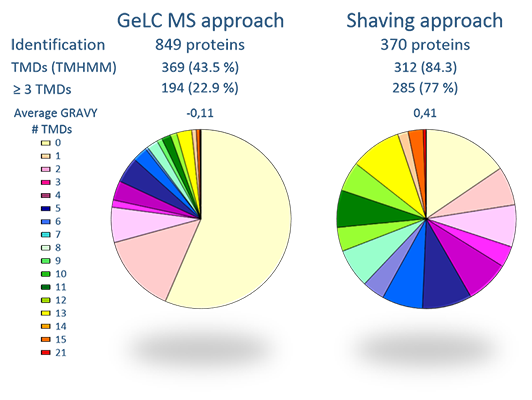
Development of enrichment schemes for bacterial membranes
Although in one third of the genome of most microorganisms predicted membrane proteins are encoded, commonly these particular proteins are underrepresented in proteome studies. The reason for this shortcoming is mainly connected to their hydrophobicity, that results in a rather incompatibility with common biochemical buffers, and in physicochemical constraints of 2D gel-based proteomic techniques. To be able to get the membrane proteome of microorganisms accessible, we have implemented and developed protocols that enrich as well as integral membrane proteins and membrane associated proteins in proteome samples and allow for 2D gel-free LC-MS/MS analysis of different membrane protein fractions.
- Wolff et al. PMID: 18460691.
- Dreisbach et al. PMID: 18491319.
- Wolff et al. PMID: 18460691.
- Hahne et al. PMID: 18763711.
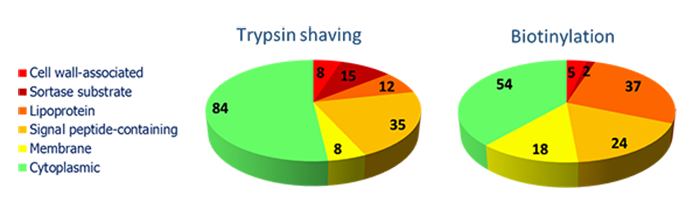
Analysis of the bacterial surface
On the bacterial surface a number of proteins with different physico-chemical properties can be observed: membrane proteins, lipoproteins and proteins attached covalently or non-covalently to the cell wall. We have developed several protocols for particular proteomic analyses, e.g. for specific labeling of these surface exposed proteins by biotinylation, or the direct enzymatic processing aimed at such protein domains exposed on surface.
- Hempel et al. PMID: 20108986.
- Dreisbach et al. PMID: 21674804.