Application of proteomics in biological and biotechnological research
Our research opens a broad variety of applications in several biological fields, thus applied proteomics could be addressed for different scientific questions in general, physiological, environmental or infection related microbiology. Therefore our workflows have been widely used in multiple studies, some examples are:
Biology of infection and stress physiology
The fast adaptation of pathogenic microbes to host defenses or new antimicrobial therapies makes it necessary to develop novel therapeutic strategies to combat such infections. Pre-requisite is a better understanding of these processes of bacterial adaptation which are often anchored deeply in fundamental biology (evolutionary mechanisms, horizontal gene transfer). As comparative proteome expression analyses give qualitatively improved insights into these adaptational networks, we are involved in numerous physiological studies. Some examples are:
- Maaβ et al. PMID: 24878497.
- Huja et al. PMID: 25118243.
- Wenzel et al. PMID: 24706874.
- Hoffmann et al. PMID: 24373249.
- Pribyl et al. PMID: 24387739.
- Lappann et al. PMID: 23893116.

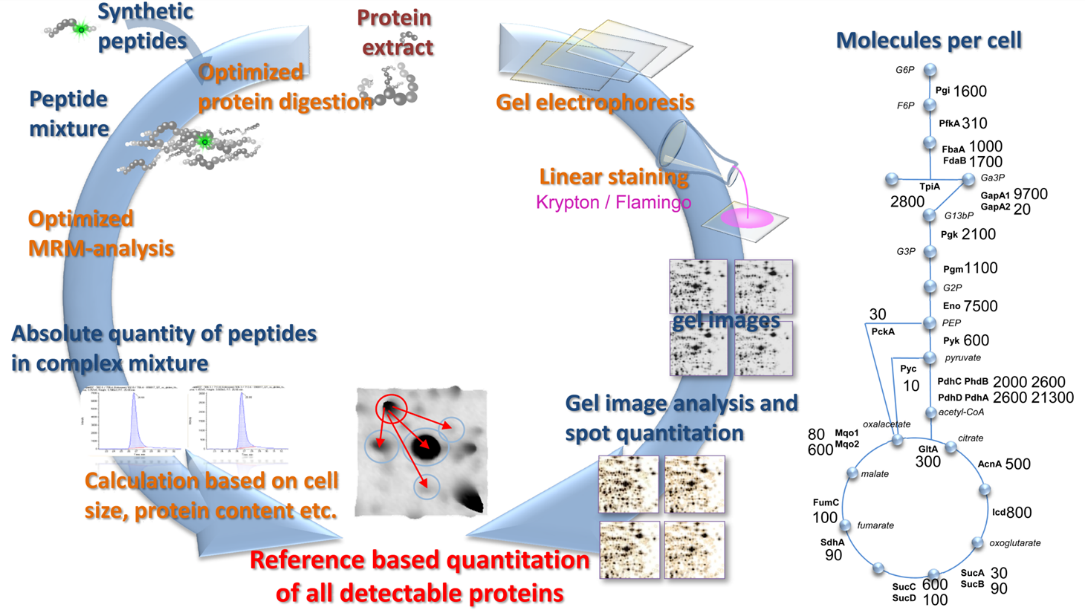
Absolute protein quantification in systems biology
In systems biology, the generation of interdependent data requires the integration of large scale ‘omics’ studies each yielding absolute data for different levels of cellular components like DNA, RNA, proteins or metabolites. Although each single ‘omics’ level data leads to unique information, the analysis of the proteome is particularly valuable due to the nature of proteins as the real effectors of cellular metabolism (enzymes and structural determinants) throughout all kingdoms of life. For generation of absolute data in systems biology approaches we rely on three different workflows:
- Determination of absolute quantities of single proteins in complex mixtures by AQUA and targeted proteomics
- 2D Gel based absolute quantification combining AQUA and state-of-the-art 2D-PAGE
- Absolute quantification based on LC-MS techniques (TOP3 on data independent MS-platforms)
- Muntel et al. PMID: 24696501.
- Maaβ et al. PMID: 24878497.
- Buescher et al. PMID: 22383848.
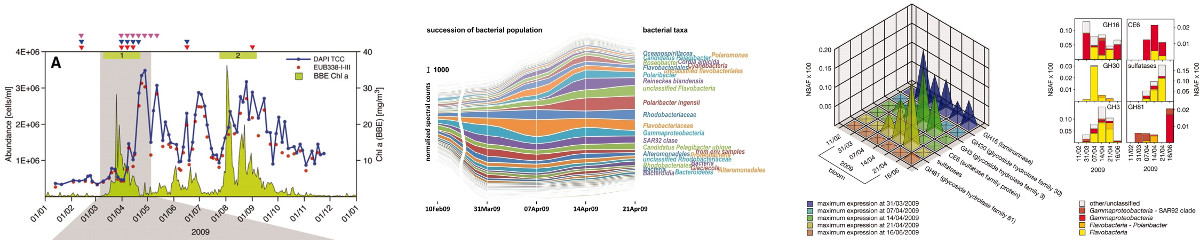
Environmental Metaproteomics
Elucidation of processes in nature has driven scientists ever since – and has gained even more attention in recent times with new technologies at hand allowing for both broad and detailed analyses of environment. Fascinating processes taking place e.g. in coastal areas, soil, microbial mats or in the human gut system that could not or not sufficiently be analyzed with technical opportunities of the pre-proteomics era. For extremely complex proteomic samples and resulting data, it is crucial to rely on state-of-the-art mass spectrometry methods as well as on highly sophisticated software capable of dealing with the information attained.
Within a network of collaborating Northern German Scientists we have succeeded in getting a glimpse into the complex bacterial successions in the North Sea at Heligoland based on multiple ‘omics techniques including proteome data.
- Xing et al. PMID: 25478683.
- Teeling et al. PMID: 22556258.